Glaucoma is the second leading cause of blindness in the world and is estimated to affect about 1 in 40 individuals over the age of 40 [1]. Despite identification of many of the pathophysiological hallmarks of glaucoma, there is no true cure for this disease. Many current therapies simply delay the onset of symptoms, and thus there is great clinical need for new treatment approaches for glaucoma patients.
One of the most well known risk factors for glaucoma is an elevated intraocular pressure (IOP) [2, 3], and consequently almost all current glaucoma therapies work via IOP reduction. Although one need not have elevated IOP (relative to the population average) in order to get glaucoma [4], the consequence of this pressure is mechanical deformation of the delicate tissues of the optic nerve head at the back of the eye, where retinal ganglion cell axons come together to form the optic nerve and pass visual information to the brain. This mechanical deformation has been hypothesized to both damage the retinal ganglion cell axons directly as well as to activate nearby astrocytes via the process of mechanotransduction, which may damage these axons [5].
We are therefore (1) investigating biomechanical strategies to control the amount of deformation that occurs in the optic nerve head under the insult of intraocular pressure, and (2) at a more fundamental level, seeking to identify the genes that control scleral stiffness.
(1) Altering the Stiffness of the Sclera to Prevent Vision Loss in Glaucoma (Bailey Hannon)
We hypothesize that if we can reduce the mechanical strain in this tissue, we may be able to protect the eye from vision loss in glaucoma. If successful, this biomechanical approach could complement current IOP-lowering therapies.
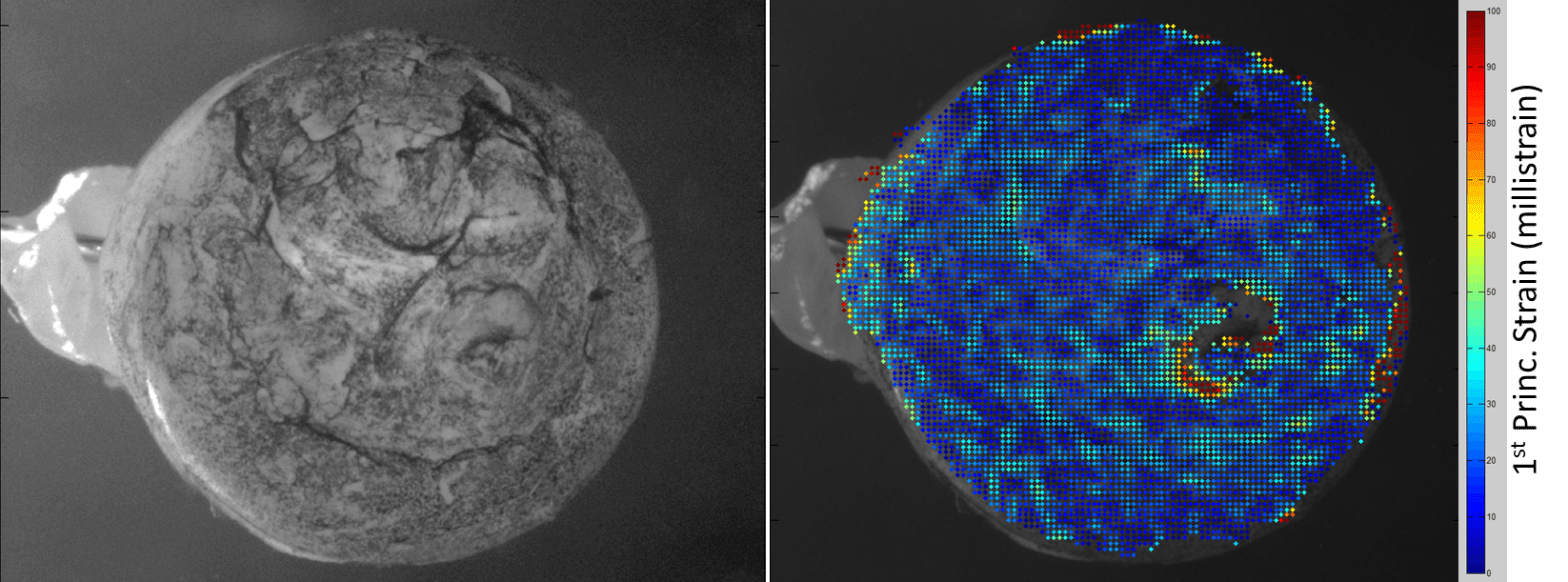
Our lab uses the magnetic microbead model of intraocular hypertension induced in rats to study the progress of this disease [6]. We can monitor their IOP using a hand-held device called a tonometer, and we can administer a rodent “eye exam” using the optomoter response as well as traditional electroretinography. We can also mechanically interrogate the eye using an optical technique called digital image correlation (Fig. 1).
Figure 1: Digital image correlation of the posterior portion of a rat eye speckled with ink as a surface tracker (left) reveals the location and magnitude of mechanical strains throughout this region (right) as IOP is manually elevated.
(2) Identifying Genes Controling Scleral Stiffness Using the BXD Mouse Family (Liz Boazak)
Successful identification of genes influencing scleral stiffness will aid in the understanding of glaucoma pathogenesis, the identification of novel therapeutic targets, the identification of at-risk patients, and the possible development of a new mouse model for glaucoma research. Ocular compliance, which can be measured using the iPerfusion system [7], indirectly reflects scleral stiffness. Direct tensile testing of mouse sclera mechanical properties is limited by tissue sample size. Furthermore, compliance measurements reflect tissue mechanical behavior in its native geometry. We expect that the genomic loci associated with ocular compliance and scleral stiffness significantly overlap. The identification of said genomic loci is possible using the BXD mouse set, which is comprised of over 100 inbred substrains with fully mapped genomes, and parent strains with fully sequenced genomes. As such, it is a powerful tool for quantitative trait locus (QTL) analysis [8], and the identification of candidate genes within identified QTLs.
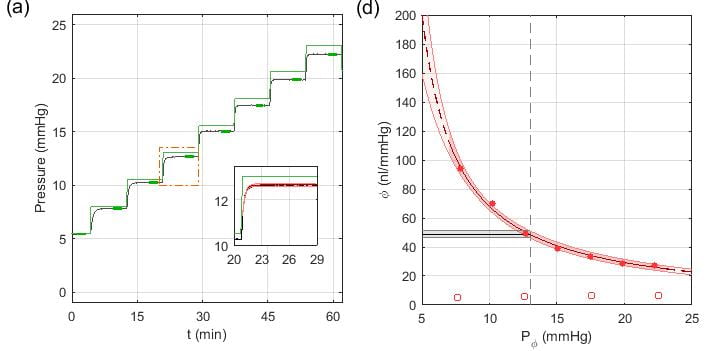
Preliminary data indicates that differences in ocular compliance can be detected between BXD mouse strain, yielding suggestive peaks in the initial QTL analysis. We have recently developed a new data analysis approach for improved precision in measuring ocular compliance (Fig 2), in which the pressure response trace to an applied increase in IOP is compared to an analytical solution. We anticipate that our improved measurement techniques, along with the evaluation of additional BXD strains will enable the identification of the genes which control scleral mechanical properties.
Figure 2: Ex vivo evaluation of ocular compliance in a C57Bl/6J mouse. (a) Measured IOP traces are shown in black, with the applied pressure step shown in green. Enlarged pressure step in inset shows the fitting with the Step Response method for the evaluation of ocular compliance, with the 95% confidence interval indicated by shading. (b) Ocular Compliance has a nonlinear relationship with pressure, which can be modeled using a modified version of Friedenwald’s equation. Compliance between eyes and animals is compared at a reference pressure of 13 mmHg, the average physiological IOP of C57 mice.
1. Quigley, H.A. and A.T. Broman, The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol, 2006. 90(3): p. 262-7.
2. Bengtsson, B. and A. Heijl, A long-term prospective study of risk factors for glaucomatous visual field loss in patients with ocular hypertension. J Glaucoma, 2005. 14(2): p. 135-8.
3. Leske, M.C., et al., Factors for glaucoma progression and the effect of treatment: the early manifest glaucoma trial. Arch Ophthalmol, 2003. 121(1): p. 48-56.
4. The Advanced Glaucoma Intervention Study (AGIS): 7. The relationship between control of intraocular pressure and visual field deterioration.The AGIS Investigators. Am J Ophthalmol, 2000. 130(4): p. 429-40.
5. Burgoyne, C.F., et al., The optic nerve head as a biomechanical structure: a new paradigm for understanding the role of IOP-related stress and strain in the pathophysiology of glaucomatous optic nerve head damage. Progress in Retinal and Eye Research, 2005. 24(1): p. 39-73.
6. Paulina A. Samsel, Lilian Kisiswa, Jonathan T. Erichsen, Stephen D. Cross, James E. Morgan, A Novel Method for the Induction of Experimental Glaucoma Using Magnetic Microspheres. Invest. Ophthalmol. Vis. Sci., 2011. 52(3):1671-1675.
7. Sherwood JM, Reina-Torres E, Bertrand JA, Rowe B, Overby DR, Measurement of Outflow Facility Using iPerfusion. PLoS ONE 11(3): e0150694.
8. www.genenetwork.org